GARSA workflow
/garsa/img/GARSA-pipeline.jpg
Database (MySQL) schema
/garsa/img/GARSA-tables.jpg
Architecture
The
current version of GARSA was implemented based on scripts, due to a
tight schedule imposed
by the need to obtain fast results on ongoing experiments. Next version
is
currently under development, and contemplates the use of Web services
and
parallelization techniques among others. Such development involves
three master
thesis and should be released within one year.
GARSAs architecture
is supported by Perl scripts, resulting on some coding effort to add
new tools
to its pipeline. However, its implementation design is modular, well
documented
and based on development standards like page templates. Its flexibility
has
been proven by recent extensions, which have been developed by
biologists
with basic Perl knowledge, in short time.
Requirements
Software
* OS: Linux (Fedora
Core tested)
* Servers: Apache 2.0 and MySQL 4.1 or higher
* Bioperl ("core"
and "run") 1.4 or
higher
Phylip 3.61-5 or
higher
ClustalW 1.83-2 or
higher
RBS finder
* CAP3
Glimmer 2.10 (for Yacop)
Glimmer 2.13
Interpro
3.3
NCBI Blast
Critica
1.05
Orpheus 2
Weblogo
Zcurve
1.02
Yacop
WU-Blast
Emboss 2.9.0.6 or
higher
* Phred/Phrap
Perl modules: perl-DBI, IO-String, perl-DBD-MySQL,
Mail-Mailer, GD-Graph, Spreadsheet::WriteExcel (get them via CPAN or RPMPan)
HTTP configuration:
The following options must be added to the the httpd.conf (Apache server) file:
<Directory "/var/www/html">
Options Indexes FollowSymLinks ExecCGI Multiviews
AddHandler cgi-script .cgi
AllowOverride AuthConfig
Order allow,deny
Allow from all
</Directory>
DirectoryIndex index.cgi
Blast databases (Pre-formatted or fasta
format):
NCBI
Uniprot
CDD
GeneOntology
Hardware
* 1 GB RAM or higher
* 20 GB Hard Disk available (80GB or higher is reccomended)
* 2.0 Ghz processor or higher
* Minimal requirements: GARSA is going to work
with those (*) minimal packages, but in a limited way as similarity
analyses won't be executed without NCBI-Blast and InterPro. Gene
prediction won't be executed without Critica package, and Phylogeny
without ClustalW and Phylip.
Download: Visit /garsa/ or
contact Dr. Alberto Dávila (davila AT ioc.fiocruz.br)
Licensing: GPL
For users interested
to use our servers for their own projects, we offer the option to host
those projects and provide advice and consultancy. The costs for it
will be evaluated case-by-case. Those costs are for hardware maintenance
and upgrade as well as consultancy, if needed. Users interested in this
option should contact Dr. Alberto Dávila "davila AT ioc.fiocruz.br". At
the moment, we have 3 Intel Xeon Dual Processor servers and over 500GB
of disk space available for this.
Starting a new Project
The only way to create a new project in GARSA is having super-user
privileges, either as "admin_garsa"
or "subadmin_garsa" user. The "admin_garsa" user can grant "subadmin_garsa" privileges to
several users, then they can create several new projects without the
need to be the "admin_garsa"
user.
Any of the above mentioned users should use the "Create Project" option of the "Project Administration" menu to
create a new project. GARSA ask for the following input data:
Project Name: scientific name
of the species to be studied, eg: Trypanosoma cruzi
or Drosophila melanogaster or
Plasmodium
falciparum
Project Code: code for the new
projet, has to be 2 letters, eg: TC or DM or PF.
Minimum Read Quality: Phred
minimum quality to be used in the chromatigrams, eg: 20
Minimum Lenght Size: Minimum Length (base pairs) of good
quality sequence that GARSA will accept, eg: 100
Project administrator name: name of the administrator of the
new project, eg: Joe Smith
Administrator email: eg: [email protected]
Administrator password: minimum
6 characters, a combination of letters and digits.
Project administrator login is
created by GARSA based on "Project Code",
eg: admin_TC or admin_DM or admin_PM
GARSA does not allow "admin_garsa" and "subadmin_garsa" users to
administer projects, the only function of these users is to create
projects.
Once a new project has been created, GARSA will send all the details of
the new project to the administrator email.
Project Configuration
New Library: GARSA asks for the folowing input data:
Library Name: eg: Fat tissue
or kDNA
or Salivary
gland
Library Code: eg: 001 or ABC
Library Description: EST Library 001
or GSS
Library ABC or ORESTES Library ZY9
Vector:
Choose a vector from the database or include a new one
using "New Vector Sequence"

Primers (Optional): Add any pair of primer sequences (forward and reverse) that
should be removed from your sequences.

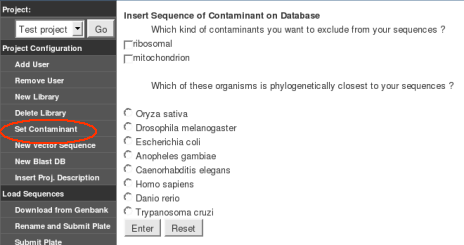
Set Contaminant (Optional): Choose
ribosomal or mithochondrial sequences that should be removed from your
sequences. The model organism more phylogentically related to the
organism to be studied by GARSA should be selected.
New Blast DB: project
administrator can load zipped multifasta files (nucleotide or
aminoacid) and format them (with formatdb) for NCBI Blast.
Load Sequences
Download from GenBank: Gene in
Genomic, EST and GSS data can be downloaded from GenBank using
scientific names, eg: Plasmodium
falciparum. GARSA shows the number of available entries, then
project administrator can decide to download the entries or not . Two
scenarios ara antecipated for the use of "Download from GenBank": a)
chromatograms are not available, then users aim to analyze data from
GenBank, b) chromatograms are available, then user aim to complement
their data with Genbank data.
Rename and Submit Plate:
chromatograms from 1 sequencing run (equivalent to a plate of 96 sloths
or less) should be copied to a single folder keeping their original
names, zipped resulting in a file as "chromats1.zip" or "reads.zip" or
"files9.zip". This zipped file is the input for GARSA. Library Code and
Plate Code should be choosen from the available options, then GARSA can
rename and upload properly the chromatograms in the zipped file.
Minimum Read Quality and Minimum Size Length can be optionally modified
here.
Submit Plate: chromatograms
from 1 sequencing run (equivalent to a plate of 96 sloths
or less) should be copied to a single folder and renamed to meet GARSA
nomenclature. In a project with DM as Project Code, JS as Lab Code, 111
as Library Code and 001 as Plate Code should contain chromatogram files
with the following names:
DMJS111001A07.g
DMJS111001C11.g
DMJS111001E08.g
Resulting Zipped File should be named: DMJS111001.zip
DMJSABC100A07.b
DMJSABC100A07.b
DMJSABC100A07.b
Resulting Zipped File should be named: DMJSABC100.zip

Only chromatograms from the same sequencing run or plate should be
zipped together, resulting in a file as "DMJS111001.zip" or
"DMJSABC100.zip". This zipped file is the input for GARSA. Minimum Read
Quality and
Minimum Size Length can be optionally modified here.
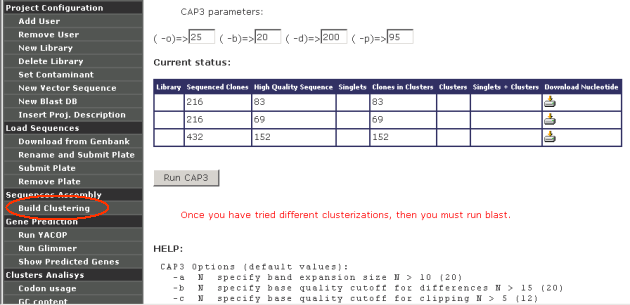
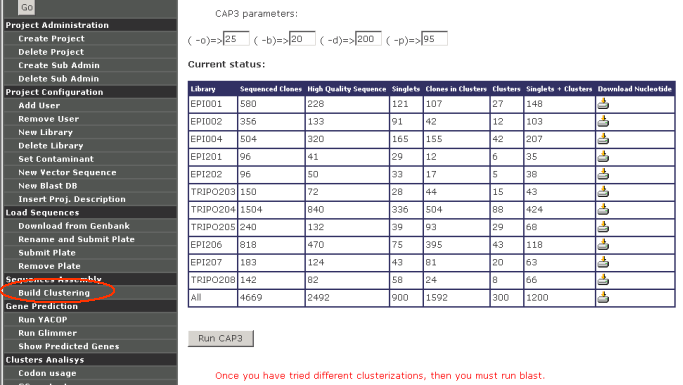
Sequence Assembly
Build clustering: Once sequences have been loaded (either in the form
of chromatograms or download from GenBank) into a given project, they
can be clustered using CAP3. The main CAP3 paramenters can be modified
several times looking for the best results.
Each time a clusterization is done, GARSA produce 1 clusterization for
each library plus 1 clusterization of all the libraries together.
Only after clusterization has been done, GARSA allows project
administrators to run Gene Prediction, Clusters Analysis and Sequence
Annotation.
Garsa shows a warning message when users try to
analyze non-clustered sequences:

Gene Prediction
GARSA can use Glimmer or the YACOP metatool (RBS, Critica, Zcurve) for
gene prediction.
Glimmer needs (complete) CDS
(multifasta format) of the organism under study or from a closely
related species to be trained.
YACOP: Critica needs a set of
nucleotide sequences from the organism under study or a closely related
species. The nucleotide sequences needed by Critica must be formatted
to be used with WU-Blast.
Sequence Annotation
Run Blast
GARSA can uses as many Blast databases as available HardDisk space. The
New Blast DB option is used to
upload and format databases. TblastX, BlastX and BlastN flavours are
active by default. However, only 2 Blast runs are allowed to happen at
the same time, in order to avoid CPU overload. E-value is configurable
at this stage.
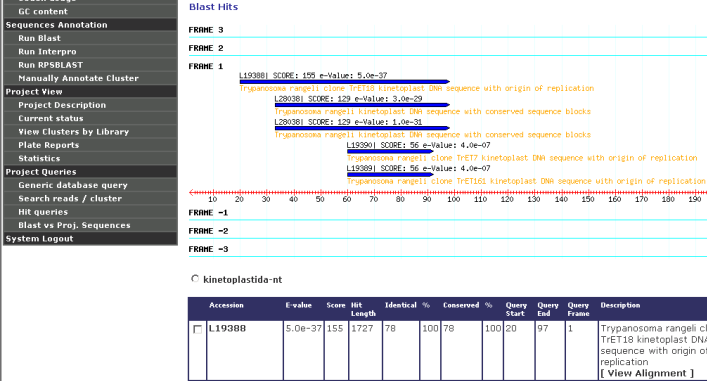
A figure showing best Blast results according to each frame is showed
aiming to help with the identification of the right frame of CDS:
Run InterPro
The current version of GARSA works with InterPro 3.2, but the new
version of GARSA (under development) will work with InterPro 4.0.
Run RPSBlast
The Conserved Domain Databases from CDD, Smart, Kog, Cog and Kegg are
available.
Notes
Users can enter comment or notes of each CLUSTER with this
option. Notes entered by user "a" cannot be deleted or modified
by user "b", then several users can work/comment the same cluster
sharing and complementing analysis.
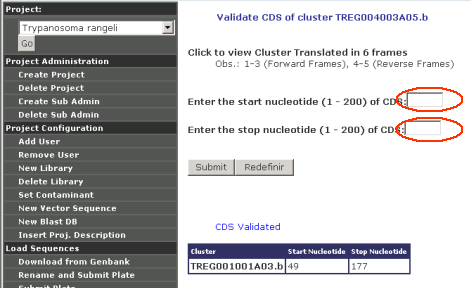
Validate CDS
When a cluster is being viewed or examined, there is always a link to
"Validate CDS":

To Validate a CDS, users need to enter the begining and end
coordinates, then Garsa translate that sequence range using the TranSeq
program of the Emboss package. Validated CDS always appear listed at
the bottom of the page:
Project queries
Generic database queries
A little console is presented, then users can query the MySQL database
using MySQL command. For security reasons, only the SELECT command is
allowed in this version.
Search Reads/Clusters
A search tool to facilitate the finding of specific reads or clusters.
Hit queries
A number of options to query the different analysis results from GARSA.
Clusters with a specific number of hits can be easily found. Clusters
with no hits can be easily found with this feature.

Blast vs Project Sequences
Garsa uses "formatdb" from the Blast package to format "Reads" and
"Clusters" to be used for WWW-Blast analysis, then any sequence can be
query against "Reads" and "Clusters" of a given project in Garsa.
Phylogeny
Users need first to clusterize
sequences using "Build Clustering" in the Sequence Assembly" menu. Most
options from the menu are only available once sequences has been
clusterized and Blast done, those results are used to help with gene
finding, alignment and phylogeny.
For Blast, "Run Blast" from the "Sequence Annotation" menu should be
used. For Logo, users should first have Blast results (after
clustering), then view results frrom a given cluster either via "View
Clusters by Library" (Project View menu) or "Search reads / cluster"
(Project Queries).


Once users are viewing the Blast results from a given cluster,
they can select one of the Blast DBs used (eg: kinetoplastida- nt)
together with their respective results:
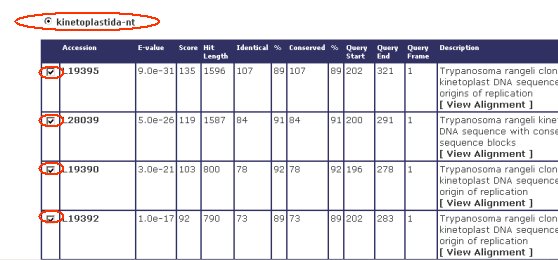
select from the bottom option what type of sequences you want to
analyze (eg: Nucleotide Sequences) then click "Run Clustal and Phylip".

After that, user will be asked for the model of substitution that
Phylip should use (eg Kimura 2- parameter). Once user have selected the
model, him/her will have a screen like this:

Acknowledgements
To
Dr. José Marcos Ribeiro (NIAID/NIH) for suggestions and sharing
his experience on EST analysis. To Dr. João Setubal (VBI and
LBI/IC/UNICAMP) for allowing us to modify the algorithm for processing
EST chromatograms. To MCT/CNPq, IAEA, CIRAD and FAPESP for financial
support. To the Open Source Community for all the valuabe
help. To the authors of the softwares/modules used in/by GARSA for
granting the academic and GPL licenses.